Main Figures from https://doi.org/10.1128/JVI.01987-13.
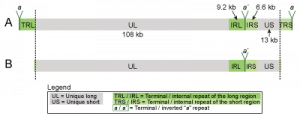
Figure 1 – Trimmed format of HSV-1 genomes
The complete HSV-1 genome includes two unique regions and two sets of large inverted repeats.
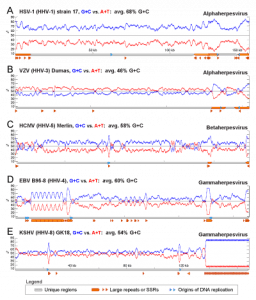
Figure 2 – G+C vs. A+T distribution spikes in herpesvirus repeat regions
Nucleotide compositional bias towards G+C residues in repeat regions of herpesvirus genomes.
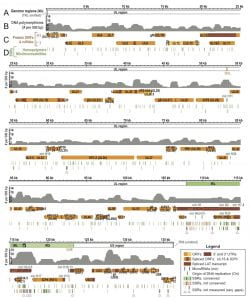
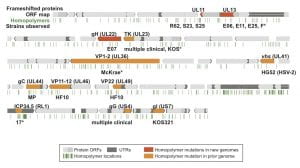
Figure 3 – Variability and conservation in the HSV-1 genome
Overview of the HSV-1 genome depicting coding regions, noncoding features, polymorphisms, and SSRs.
Supplemental Figures from https://doi.org/10.1128/JVI.01987-13.
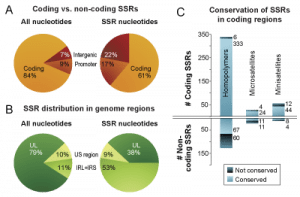
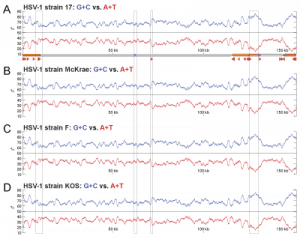
Figure S1 – GC vs. AT distribution in multiple HSV-1 strains
G+C-richness peaks in terminal and inverted repeats, in a pattern common across HSV-1 strains.
Additional Figures
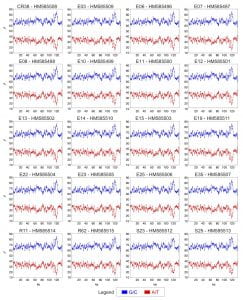
Figure A1 – GC vs. AT distribution in twenty HSV-1 genomes
Nucleotide abundance of G+C vs. A+T for 20 new HSV-1 genomes. Line graph of G+C vs. A+T distribution in the twenty newly sequenced HSV-1 genomes reveals a pattern matching other HSV-1 strains (Figure 2). The trimming of terminal copies of the large repeats (Figure 1) removes the outermost spikes of G+C-richness seen in Figure 2A.