Research Summary / Jump to: Gallery or Data websites
Research Interests
Comparative genomics of herpesviruses, neurovirology, pathogen genetic & phenotypic diversity, neuron-virus relationships
Research Summary
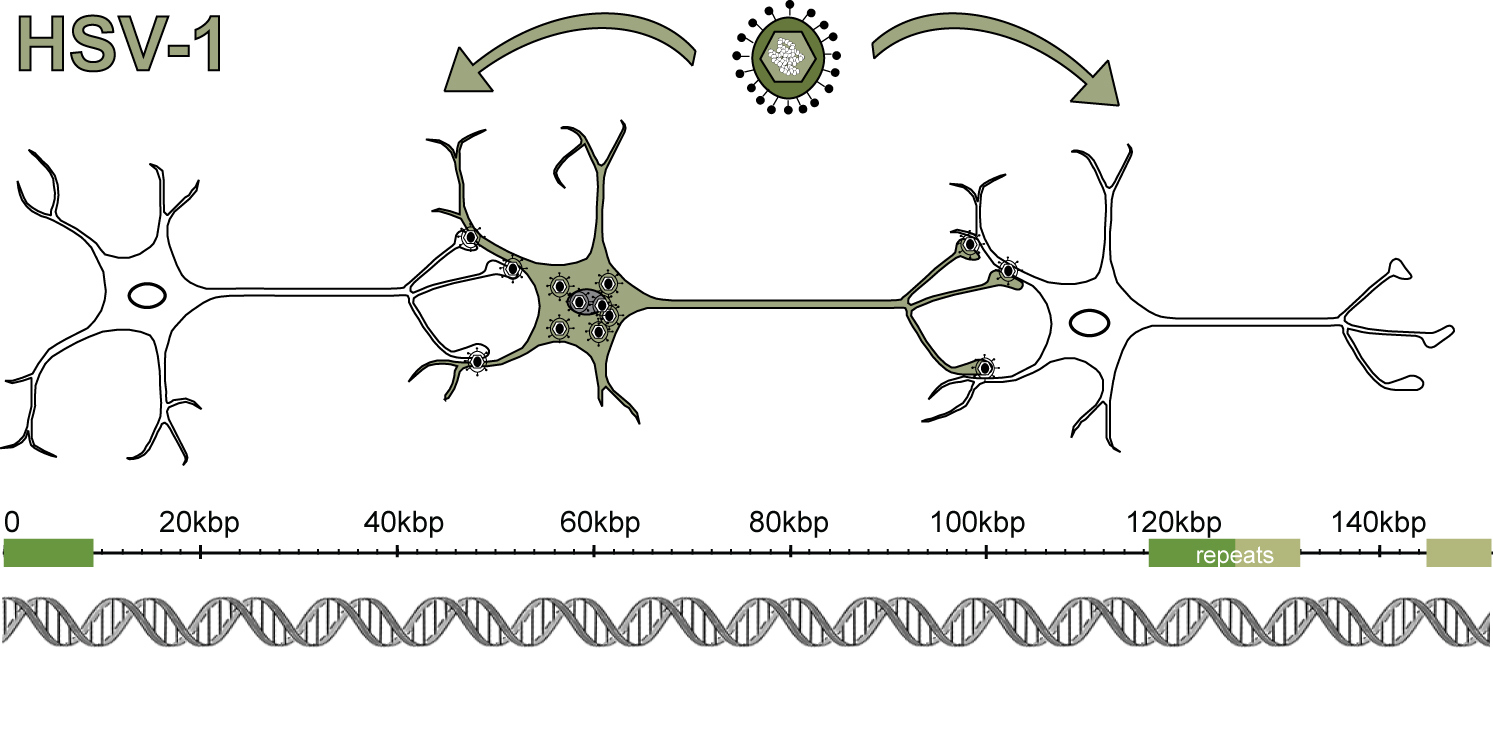
A variety of viruses infect the human nervous system, often with severe consequences. While vaccines have largely defeated the paralysis caused by polio, other viruses such as rabies, West Nile virus, and herpes simplex virus (HSV) continue to cause neurological infections that require clinical intervention. More than 65% of adults in the United States carry HSV, whether they know it or not. HSV causes recurrent genital and oral lesions (e.g. cold sores), with different levels of lesion severity and rates of recurrence observed in different individuals. HSV can also spread and cause symptoms elsewhere, such as in the eyes (e.g., ocular keratitis), brain (e.g., encephalitis or meningitis), or liver (e.g., hepatitis). In newborns, more than half of all HSV infections disseminate into the brain and/or the body, causing much more severe outcomes than occurs in adults. Once infected, humans have a permanent relationship with this virus, because of this virus’ unique ability to establish lifelong latency in neurons. The consequences of HSV latency for the neurons that harbor this pathogen are not well understood. Our laboratory aims to address how the genetic features of different HSV infections — and the specific aspects of infected hosts — impact the outcomes of these infections. To do this, we use a combination of virology, neurobiology, next generation sequencing technologies, and bioinformatics.
Viral variation and neurovirulence
We have known for a long time that strains of HSV-1 are genetically variable. Deep genome-sequencing approaches now allow us to fully define these differences and begin to understand how they affect phenotype. In collaboration with our clinician-scientist colleagues, we have begun to define the viral genomic diversity found in human infections. We have found that human HSV-1 infections can be genetically narrow (i.e., mostly homogeneous) or quite diverse (i.e., heterogeneous), and that viral genetic diversity can be passed on to others. Using animal models where it is possible to control for host genetics, prior researchers have determined that HSV-1 strains can differ in their level of neurovirulence — i.e., their ability to cause disease in the central nervous system. We are working to connect data such as these to the range of clinical disease outcomes observed in humans, for instance in the range of mild vs. severe outcomes observed in infected newborns. Previous tools to map differences between HSV strains included restriction digest mapping or candidate-gene approaches. Now we can readily sequence the full genomes of many strains and use bioinformatics to associate variations with observed phenotypes. These approaches produce hypotheses that we can then test experimentally. We anticipate that viral population diversity contributes to the symptoms and progression of HSV disease in humans.
Viral evolution and drug resistance
Humans infected with HSV-1 carry a “population” of viruses, which is generally composed of a dominant viral genotype and an array of minor viral variants (i.e., less common viral genetic variants that also exist in the infected host). Minor variants in the viral population can contribute to the fitness and/or the evolution of the viral population. These variants may undergo rapid selection if they confer a fitness benefit, e.g., a drug resistance allele may rapidly become the dominant genotype in the presence of antiviral drug selective pressure. Other missense mutations or changes in regulatory sequences may impact viral fitness, or contribute to viral recombination. We use experimental evolution to model the changes that arise in HSV genomes over time, and under different selective pressures. We anticipate that these studies will help reveal the rate at which viral genetic diversity arises, and how this diversity influences observed phenotypes such as antiviral drug resistance.
Neuronal consequences of infection
Because HSV sets up a lifelong latency in neurons, it is critical that we understand how this long-term viral presence affects human neurons and their neighboring cells. In lab experiments, HSV infection of neurons has been associated with changes in neuropeptide release, neuronal firing rates, axon remodeling, and interferon production. We and others have established cell-based methods that use human neurons to model HSV-1 infection of neurons in controlled conditions in the lab. Using these models, we have shown that different strains of HSV-1 have different programs of gene expression in human neurons, which contributes to differences in their observed phenotypes. Features unique to neurons may present novel targets to block the progression of infection. We address the neurobiology of HSV infection using a combination of neuronal cultures, in vivo models of infection, and high-throughput measurements of the host response.